132 DNA Structure and Sequencing
Learning Objectives
By the end of this section, you will be able to do the following:
- Describe the structure of DNA
- Explain the Sanger method of DNA sequencing
- Discuss the similarities and differences between eukaryotic and prokaryotic DNA
The building blocks of DNA are nucleotides. The important components of the nucleotide are a nitrogenous (nitrogen-bearing) base, a 5-carbon sugar (pentose), and a phosphate group (Figure 14.5). The nucleotide is named depending on the nitrogenous base. The nitrogenous base can be a purine such as adenine (A) and guanine (G), or a pyrimidine such as cytosine (C) and thymine (T).
Visual Connection
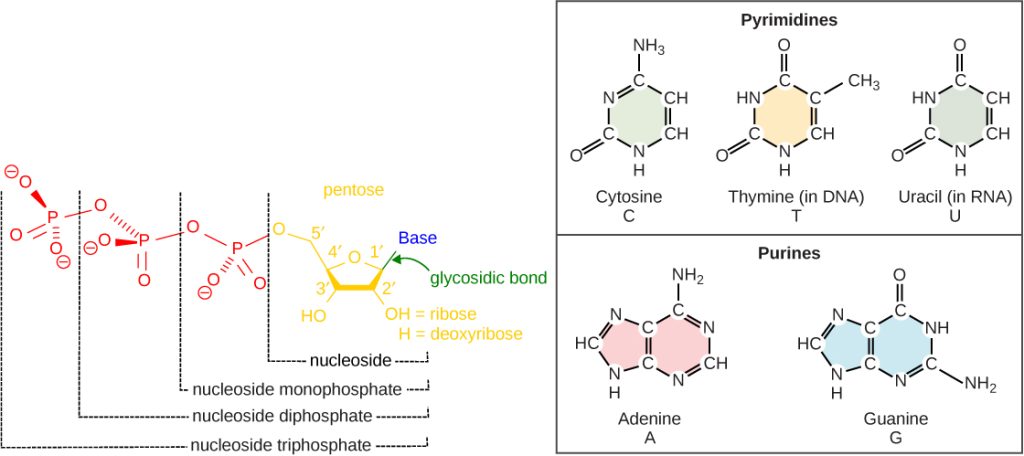
The purines have a double ring structure with a six-membered ring fused to a five-membered ring. Pyrimidines are smaller in size; they have a single six-membered ring structure.
The sugar is deoxyribose in DNA and ribose in RNA. The carbon atoms of the five-carbon sugar are numbered 1′, 2′, 3′, 4′, and 5′ (1′ is read as “one prime”). The phosphate, which makes DNA and RNA acidic, is connected to the 5′ carbon of the sugar by the formation of an ester linkage between phosphoric acid and the 5′-OH group (an ester is an acid + an alcohol). In DNA nucleotides, the 3′ carbon of the sugar deoxyribose is attached to a hydroxyl (OH) group. In RNA nucleotides, the 2′ carbon of the sugar ribose also contains a hydroxyl group. The base is attached to the 1’carbon of the sugar.
The nucleotides combine with each other to produce phosphodiester bonds. The phosphate residue attached to the 5′ carbon of the sugar of one nucleotide forms a second ester linkage with the hydroxyl group of the 3′ carbon of the sugar of the next nucleotide, thereby forming a 5′-3′ phosphodiester bond. In a polynucleotide, one end of the chain has a free 5′ phosphate, and the other end has a free 3′-OH. These are called the 5′ and 3′ ends of the chain.
Rosalind Franklin joined the scientists at the Medical Research Unit, King’s College when John Randall recruited her to work on the structure of DNA. DNA (deoxyribonucleic acid) was originally discovered in 1898 by Johann Miescher, and it was known that it was a key to genetics. But it was not until the middle of the 20th century when scientific methods had developed to where the actual structure of the molecule could be discovered, and Rosalind Franklin’s work was key to that methodology.
Rosalind Franklin worked on the DNA molecule from 1951 until 1953. Using x-ray crystallography, she took photographs of the B version of the molecule. A co-worker with whom Franklin did not have a good working relationship, Maurice H.F. Wilkins, showed Franklin’s photographs of DNA to James Watson—without Franklin’s permission. Watson and his research partner Francis Crick were working independently on the structure of DNA, and Watson realized that these photographs were the scientific evidence they needed to prove that the DNA molecule was a double-stranded helix.


In the 1950s, Francis Crick and James Watson worked together to determine the structure of DNA at the University of Cambridge, England. Other scientists like Linus Pauling and Maurice Wilkins were also actively exploring this field. Pauling previously had discovered the secondary structure of proteins using X-ray crystallography. In Wilkins’ lab, researcher Rosalind Franklin was using X-ray diffraction methods to understand the structure of DNA. Watson and Crick were able to piece together the puzzle of the DNA molecule on the basis of Franklin’s data because Crick had also studied X-ray diffraction (Figure 14.6). In 1962, James Watson, Francis Crick, and Maurice Wilkins were awarded the Nobel Prize in Medicine. Unfortunately, by then Franklin had died, and Nobel prizes are not awarded posthumously.
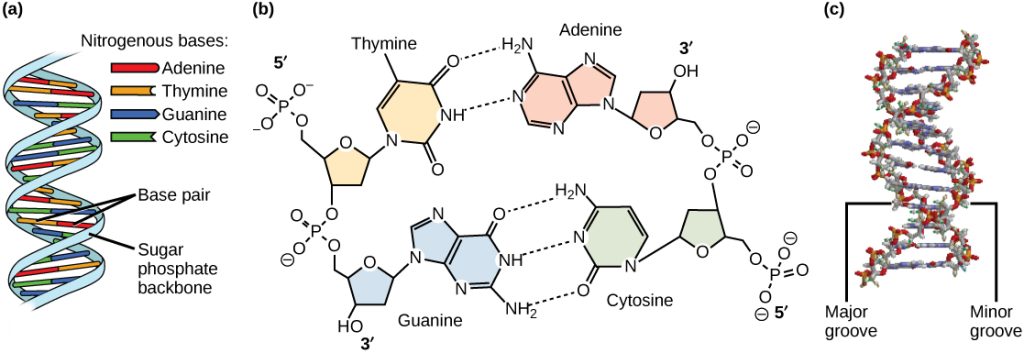
Watson and Crick proposed that DNA is made up of two strands that are twisted around each other to form a right-handed helix. Base pairing takes place between a purine and pyrimidine on opposite strands, so that A pairs with T, and G pairs with C (suggested by Chargaff’s Rules). Thus, adenine and thymine are complementary base pairs, and cytosine and guanine are also complementary base pairs. The base pairs are stabilized by hydrogen bonds: adenine and thymine form two hydrogen bonds and cytosine and guanine form three hydrogen bonds. The two strands are anti-parallel in nature; that is, the 3′ end of one strand faces the 5′ end of the other strand. The sugar and phosphate of the nucleotides form the backbone of the structure, whereas the nitrogenous bases are stacked inside, like the rungs of a ladder. Each base pair is separated from the next base pair by a distance of 0.34 nm, and each turn of the helix measures 3.4 nm. Therefore, 10 base pairs are present per turn of the helix. The diameter of the DNA double-helix is 2 nm, and it is uniform throughout. Only the pairing between a purine and pyrimidine and the antiparallel orientation of the two DNA strands can explain the uniform diameter. The twisting of the two strands around each other results in the formation of uniformly spaced major and minor grooves (Figure 14.7).
DNA Sequencing Techniques
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, guanine, cytosine, and thymine. The advent of rapid DNA sequencing methods has greatly accelerated biological and medical research and discovery. Until the 1990s, the sequencing of DNA (reading the sequence of DNA) was a relatively expensive and long process. Using radiolabeled nucleotides also compounded the problem through safety concerns. With currently available technology and automated machines, the process is cheaper, safer, and can be completed in a matter of hours. Fred Sanger developed the sequencing method used for the human genome sequencing project, which is widely used today (Figure 14.8).
Link to Learning
Visit this site to watch a video explaining the DNA sequence-reading technique that resulted from Sanger’s work. These videos provide more information on DNA translation: DNA Translation: Initiation Phase & Ribosome FormationDNA Translation: mRNA to Protein, and tRNA’s RoleDNA as Genetic material: Avery-MacLeod-McCarty experiment
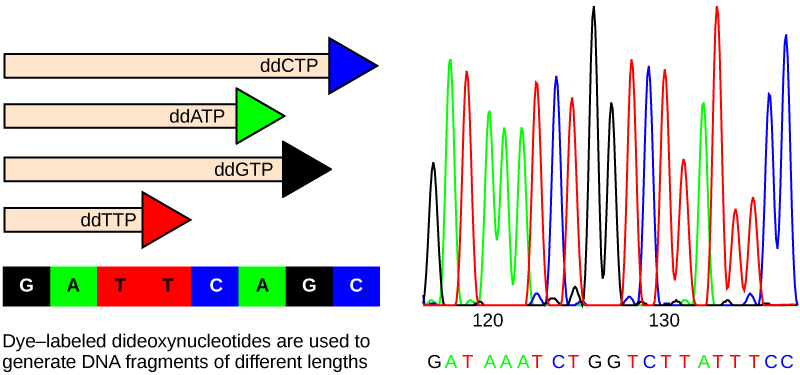
The DNA sample to be sequenced is denatured (separated into two strands by heating it to high temperatures). The DNA is divided into four tubes in which a primer, DNA polymerase, and all four nucleoside triphosphates (A, T, G, and C) are added. In addition, limited quantities of one of the four dideoxynucleoside triphosphates (ddCTP, ddATP, ddGTP, and ddTTP) are added to each tube respectively. The tubes are labeled as A, T, G, and C according to the ddNTP added. For detection purposes, each of the four dideoxynucleotides carries a different fluorescent label. Chain elongation continues until a fluorescent dideoxy nucleotide is incorporated, after which no further elongation takes place. After the reaction is over, electrophoresis is performed. Even a difference in length of a single base can be detected. The sequence is read from a laser scanner that detects the fluorescent marker of each fragment. For his work on DNA sequencing, Sanger received a Nobel Prize in Chemistry in 1980.
Link to Learning
Sanger’s genome sequencing has led to a race to sequence human genomes at rapid speed and low cost. Learn more by viewing the animation here.
Gel electrophoresis is a technique used to separate DNA fragments of different sizes. Usually the gel is made of a chemical called agarose (a polysaccharide polymer extracted from seaweed that is high in galactose residues). Agarose powder is added to a buffer and heated. After cooling, the gel solution is poured into a casting tray. Once the gel has solidified, the DNA is loaded on the gel and electric current is applied. The DNA has a net negative charge and moves from the negative electrode toward the positive electrode. The electric current is applied for sufficient time to let the DNA separate according to size; the smallest fragments will be farthest from the well (where the DNA was loaded), and the heavier molecular weight fragments will be closest to the well. Once the DNA is separated, the gel is stained with a DNA-specific dye for viewing it (Figure 14.9).
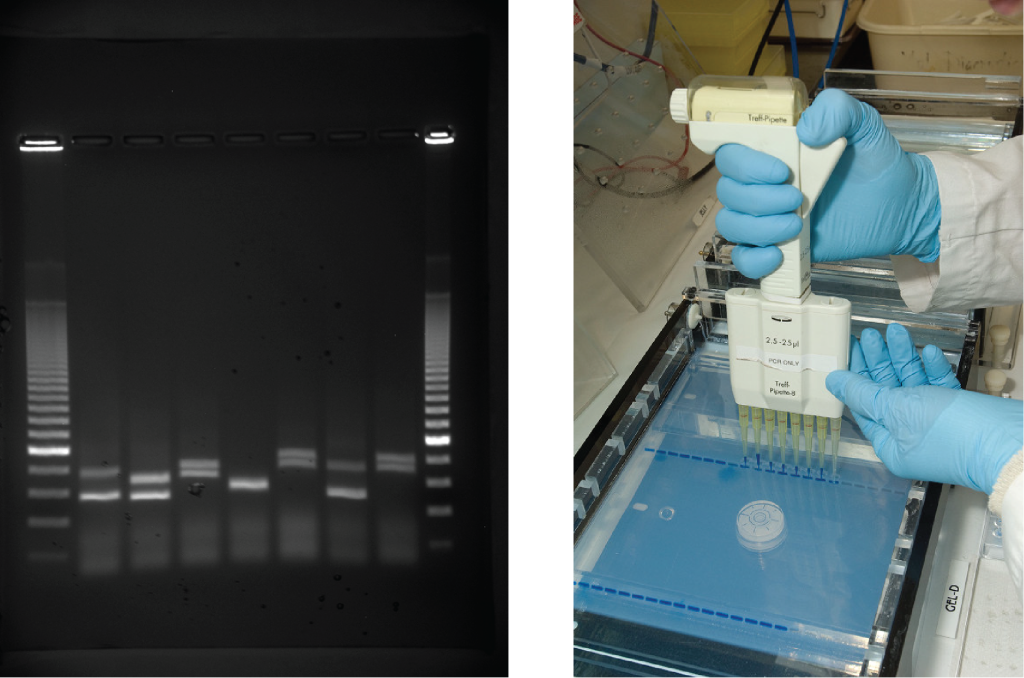
Evolution Connection
Neanderthal Genome: How Are We Related? The first draft sequence of the Neanderthal genome was recently published by Richard E. Green et al. in 2010.1 Neanderthals are the closest ancestors of present-day humans. They were known to have lived in Europe and Western Asia (and now, perhaps, in Northern Africa) before they disappeared from fossil records approximately 30,000 years ago. Green’s team studied almost 40,000-year-old fossil remains that were selected from sites across the world. Extremely sophisticated means of sample preparation and DNA sequencing were employed because of the fragile nature of the bones and heavy microbial contamination. In their study, the scientists were able to sequence some four billion base pairs. The Neanderthal sequence was compared with that of present-day humans from across the world. After comparing the sequences, the researchers found that the Neanderthal genome had 2 to 3 percent greater similarity to people living outside Africa than to people in Africa. While current theories have suggested that all present-day humans can be traced to a small ancestral population in Africa, the data from the Neanderthal genome suggest some interbreeding between Neanderthals and early modern humans.
Link to Learning
Watch Svante Pääbo’s talk explaining the Neanderthal genome research at the 2011 annual TED (Technology, Entertainment, Design) conference. This review video demonstrates how DNA is packaged.
DNA Packaging in Cells
Visual Connection
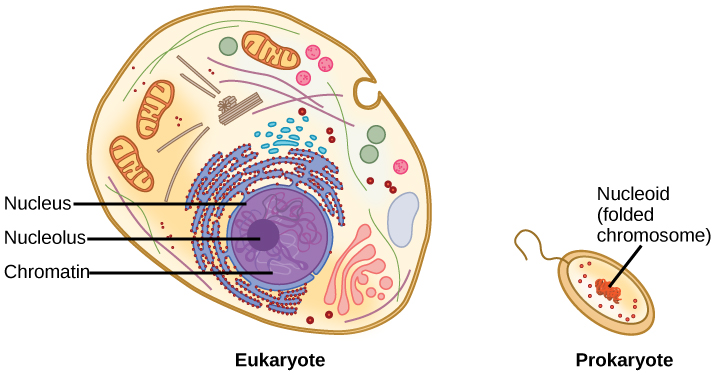
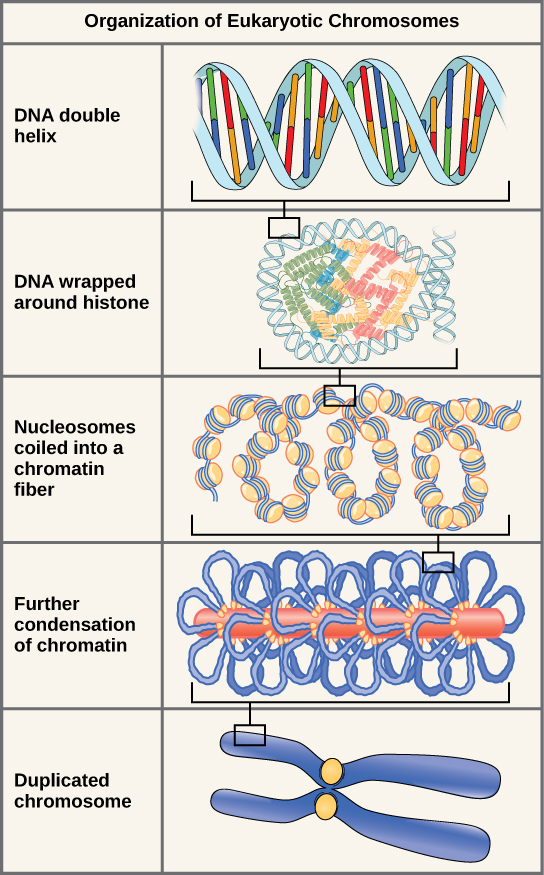
The size of the genome in one of the most well-studied prokaryotes, E.coli, is 4.6 million base pairs (approximately 1.1 mm, if cut and stretched out). So how does this fit inside a small bacterial cell? The DNA is twisted by what is known as supercoiling. Supercoiling suggests that DNA is either “under-wound” (less than one turn of the helix per 10 base pairs) or “over-wound” (more than 1 turn per 10 base pairs) from its normal relaxed state. Some proteins are known to be involved in the supercoiling; other proteins and enzymes such as DNA gyrase help in maintaining the supercoiled structure. Eukaryotes, whose chromosomes each consist of a linear DNA molecule, employ a different type of packing strategy to fit their DNA inside the nucleus (Figure 14.11). At the most basic level, DNA is wrapped around proteins known as histones to form structures called nucleosomes. The histones are evolutionarily conserved proteins that are rich in basic amino acids and form an octamer composed of two molecules of each of four different histones. Their composition and properties are important to understanding gene expression, and were partially uncovered based on research by Marie M. Daly and Alfred E. Mirsky in the early 1950s. The DNA (remember, it is negatively charged because of the phosphate groups) is wrapped tightly around the histone core. This nucleosome is linked to the next one with the help of a linker DNA. This is also known as the “beads on a string” structure. With the help of a fifth histone, a string of nucleosomes is further compacted into a 30-nm fiber, which is the diameter of the structure. Metaphase chromosomes are even further condensed by association with scaffolding proteins. At the metaphase stage, the chromosomes are at their most compact, approximately 700 nm in width. In interphase, eukaryotic chromosomes have two distinct regions that can be distinguished by staining. The tightly packaged region is known as heterochromatin, and the less dense region is known as euchromatin. Heterochromatin usually contains genes that are not expressed, and is found in the regions of the centromere and telomeres. The euchromatin usually contains genes that are transcribed, with DNA packaged around nucleosomes but not further compacted.
Footnotes
- 1 Richard E. Green et al., “A Draft Sequence of the Neandertal Genome,” Science 328 (2010): 710-22.

